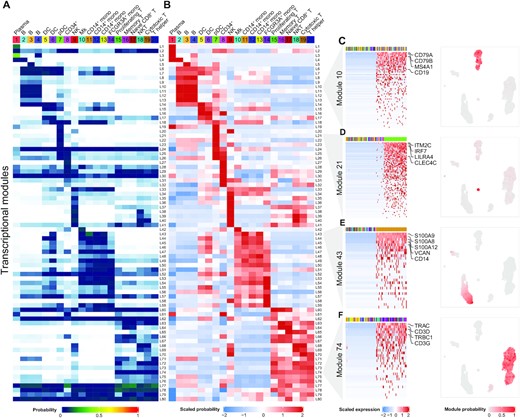
Single-cell RNA-seq (scRNA-seq) has emerged as a powerful technique to quantify gene expression in individual cells and to elucidate the molecular and cellular building blocks of complex tissues. We developed a novel Bayesian hierarchical model called Cellular Latent Dirichlet Allocation (Celda) to perform co-clustering of genes into transcriptional modules and cells into subpopulations. Celda can quantify the probabilistic contribution of each gene to each module, each module to each cell population and each cell population to each sample. In a peripheral blood mononuclear cell dataset, Celda identified a subpopulation of proliferating T cells and a plasma cell which were missed by two other common single-cell workflows. Celda also identified transcriptional modules that could be used to characterize unique and shared biological programs across cell types. Finally, Celda outperformed other approaches for clustering genes into modules on simulated data. Celda presents a novel method for characterizing transcriptional programs and cellular heterogeneity in scRNA-seq data.
Wang, Z. et al. Celda: a Bayesian model to perform co-clustering of genes into modules and cells into subpopulations using single-cell RNA-seq data. NAR Genomics Bioinforma. 4, (2022). https://doi.org/10.1093/nargab/lqac066