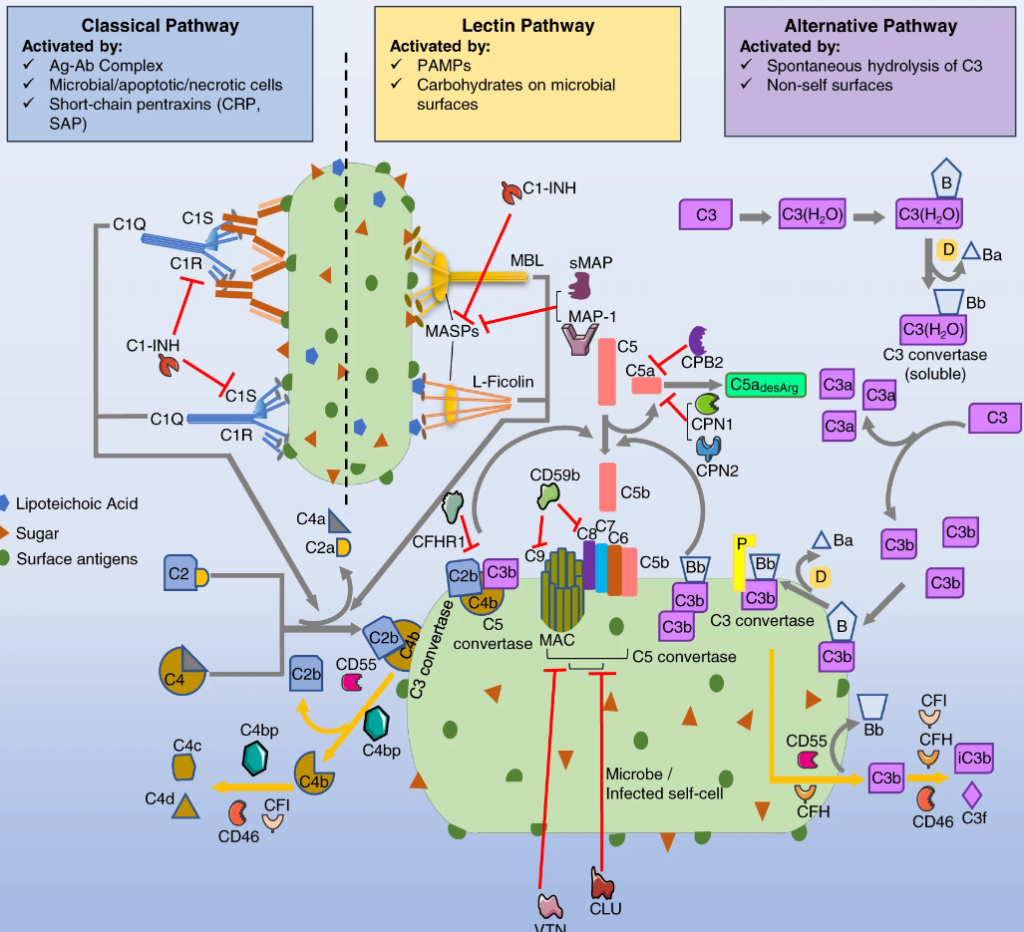
To understand functional duality of the complement system in host defense and lung injury, a more comprehensive view of its localized production in the lung, and the impact of age on complement production are essential. Here, we explored the expression of complement genes through computational analysis of preexisting single cell RNA sequencing data from lung transcriptomes of healthy young (3 months) and old C57BL/6 mice (24 months), and humans. We characterized the distribution of 48 complement genes. Across 28 distinct immune and non-immune cell types in mice, mesothelial cells expressed the greatest number of complement genes (e.g., C1ra, C2, C3), and regulators (e.g., Serping1, Cfh). C5 was abundant in type II alveolar epithelial cells and C1q in interstitial lung macrophages. There were only moderate differences in gene expression between young and old mice. Among 57 human lung cell types, mesothelial cells showed abundant complement expression. A few differences in gene expression (e.g., FCN1, CFI, C6, C7) were also evident between mice and human lung cells. Our findings present a novel perspective on the expression patterns of complement genes in normal lungs. These findings highlight the potential functions of complement in tissue-specific homeostasis and immunity and may foster a mechanistic understanding of its role in lung health and disease.