Shiyi Yang, Sean E. Corbett, Yusuke Koga, Zhe Wang, W. Evan Johnson, Masanao Yajima, Joshua D. Campbell.
Abstract
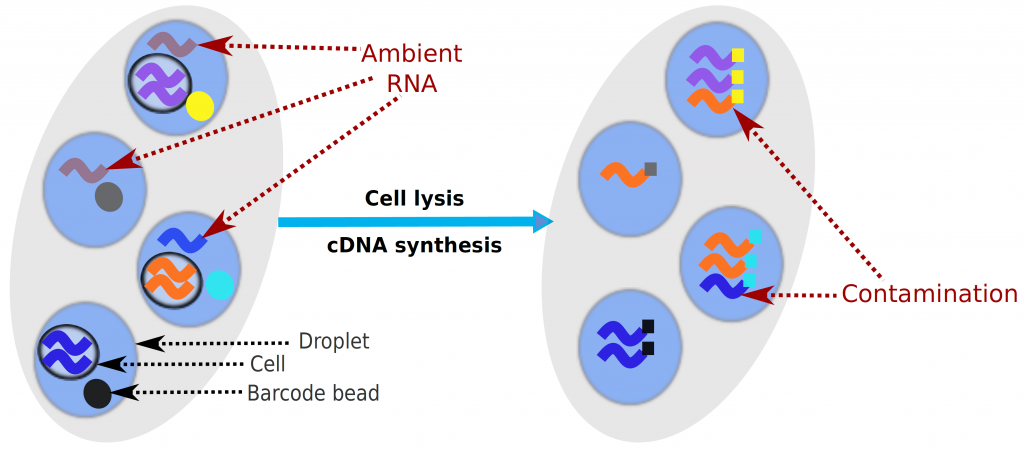
Droplet-based microfluidic devices have become widely used to perform single-cell RNA sequencing (scRNA-seq). However, ambient RNA present in the cell suspension can be aberrantly counted along with a cell’s native mRNA and result in cross-contamination of transcripts between different cell populations. DecontX is a novel Bayesian method to estimate and remove contamination in individual cells. DecontX accurately predicts contamination levels in a mouse-human mixture dataset and removes aberrant expression of marker genes in PBMC datasets. We also compare the contamination levels between four different scRNA-seq protocols. Overall, DecontX can be incorporated into scRNA-seq workflows to improve downstream analyses.
Keywords: Bayesian mixture model; Decontamination; Single cell; scRNA-seq.