Uses the celda_C model to cluster cells into
population for range of possible K's. The cell population labels of the
previous "K-1" model are used as the initial values in the current model
with K cell populations. The best split of an existing cell population is
found to create the K-th cluster. This procedure is much faster than
randomly initializing each model with a different K. If module labels for
each feature are given in 'yInit', the celda_CG model will be used to
split cell populations based on those modules instead of individual
features. Module labels will also be updated during sampling and thus
may end up slightly different than yInit.
recursiveSplitCell(
x,
useAssay = "counts",
altExpName = "featureSubset",
sampleLabel = NULL,
initialK = 5,
maxK = 25,
tempL = NULL,
yInit = NULL,
alpha = 1,
beta = 1,
delta = 1,
gamma = 1,
minCell = 3,
reorder = TRUE,
seed = 12345,
perplexity = TRUE,
doResampling = FALSE,
numResample = 5,
logfile = NULL,
verbose = TRUE
)
# S4 method for SingleCellExperiment
recursiveSplitCell(
x,
useAssay = "counts",
altExpName = "featureSubset",
sampleLabel = NULL,
initialK = 5,
maxK = 25,
tempL = NULL,
yInit = NULL,
alpha = 1,
beta = 1,
delta = 1,
gamma = 1,
minCell = 3,
reorder = TRUE,
seed = 12345,
perplexity = TRUE,
doResampling = FALSE,
numResample = 5,
logfile = NULL,
verbose = TRUE
)
# S4 method for matrix
recursiveSplitCell(
x,
useAssay = "counts",
altExpName = "featureSubset",
sampleLabel = NULL,
initialK = 5,
maxK = 25,
tempL = NULL,
yInit = NULL,
alpha = 1,
beta = 1,
delta = 1,
gamma = 1,
minCell = 3,
reorder = TRUE,
seed = 12345,
perplexity = TRUE,
doResampling = FALSE,
numResample = 5,
logfile = NULL,
verbose = TRUE
)Arguments
- x
A numeric matrix of counts or a SingleCellExperiment with the matrix located in the assay slot under
useAssay. Rows represent features and columns represent cells.- useAssay
A string specifying the name of the assay slot to use. Default "counts".
- altExpName
The name for the altExp slot to use. Default "featureSubset".
- sampleLabel
Vector or factor. Denotes the sample label for each cell (column) in the count matrix.
- initialK
Integer. Initial number of cell populations to try. Default
5.- maxK
Integer. Maximum number of cell populations to try. Default
25.- tempL
Integer. Number of temporary modules to identify and use in cell splitting. Only used if
yInit = NULL. Collapsing features to a relatively smaller number of modules will increase the speed of clustering and tend to produce better cell populations. This number should be larger than the number of true modules expected in the dataset. DefaultNULL.- yInit
Integer vector. Module labels for features. Cells will be clustered using the celda_CG model based on the modules specified in
yInitrather than the counts of individual features. While the features will be initialized to the module labels inyInit, the labels will be allowed to move within each new model with a different K.- alpha
Numeric. Concentration parameter for Theta. Adds a pseudocount to each cell population in each sample. Default
1.- beta
Numeric. Concentration parameter for Phi. Adds a pseudocount to each feature in each cell (if
yInitis NULL) or to each module in each cell population (ifyInitis set). Default1.- delta
Numeric. Concentration parameter for Psi. Adds a pseudocount to each feature in each module. Only used if
yInitis set. Default 1.- gamma
Numeric. Concentration parameter for Eta. Adds a pseudocount to the number of features in each module. Only used if
yInitis set. Default 1.- minCell
Integer. Only attempt to split cell populations with at least this many cells.
- reorder
Logical. Whether to reorder cell populations using hierarchical clustering after each model has been created. If FALSE, cell populations numbers will correspond to the split which created the cell populations (i.e. 'K15' was created at split 15, 'K16' was created at split 16, etc.). Default TRUE.
- seed
Integer. Passed to with_seed. For reproducibility, a default value of 12345 is used. If NULL, no calls to with_seed are made.
- perplexity
Logical. Whether to calculate perplexity for each model. If FALSE, then perplexity can be calculated later with resamplePerplexity. Default TRUE.
- doResampling
Boolean. If
TRUE, then each cell in the counts matrix will be resampled according to a multinomial distribution to introduce noise before calculating perplexity. DefaultFALSE.- numResample
Integer. The number of times to resample the counts matrix for evaluating perplexity if
doResamplingis set toTRUE. Default5.- logfile
Character. Messages will be redirected to a file named "logfile". If NULL, messages will be printed to stdout. Default NULL.
- verbose
Logical. Whether to print log messages. Default TRUE.
Value
A SingleCellExperiment object. Function parameter settings and celda model results are stored in the
metadata
"celda_grid_search" slot. The models in
the list will be of class celda_C if yInit = NULL or
celda_CG if zInit is set.
See also
recursiveSplitModule for recursive splitting of feature modules.
Examples
data(sceCeldaCG)
## Create models that range from K = 3 to K = 7 by recursively splitting
## cell populations into two to produce \link{celda_C} cell clustering models
sce <- recursiveSplitCell(sceCeldaCG, initialK = 3, maxK = 7)
#> ==================================================
#> Starting recursive cell population splitting.
#> ==================================================
#> Mon Nov 6 08:01:01 2023 .. Initializing with 3 populations
#> Mon Nov 6 08:01:01 2023 .. Current cell population 4 | logLik: -1225755.01101897
#> Mon Nov 6 08:01:01 2023 .. Current cell population 5 | logLik: -1213677.60126784
#> Mon Nov 6 08:01:01 2023 .. Current cell population 6 | logLik: -1213903.59449854
#> Mon Nov 6 08:01:01 2023 .. Current cell population 7 | logLik: -1214081.54311397
#> Mon Nov 6 08:01:01 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive cell population splitting. Total time: 0.2422531 secs
#> ==================================================
## Alternatively, first identify features modules using
## \link{recursiveSplitModule}
moduleSplit <- recursiveSplitModule(sceCeldaCG, initialL = 3, maxL = 15)
#> ==================================================
#> Starting recursive module splitting.
#> ==================================================
#> Mon Nov 6 08:01:01 2023 .. Collapsing to 100 temporary cell populations
#> Mon Nov 6 08:01:02 2023 .. Initializing with 3 modules
#> Mon Nov 6 08:01:02 2023 .. Created module 4 | logLik: -1241379.90928455
#> Mon Nov 6 08:01:02 2023 .. Created module 5 | logLik: -1235212.7977535
#> Mon Nov 6 08:01:02 2023 .. Created module 6 | logLik: -1232789.9817561
#> Mon Nov 6 08:01:02 2023 .. Created module 7 | logLik: -1227246.66090571
#> Mon Nov 6 08:01:03 2023 .. Created module 8 | logLik: -1223898.757694
#> Mon Nov 6 08:01:03 2023 .. Created module 9 | logLik: -1221848.26936098
#> Mon Nov 6 08:01:03 2023 .. Created module 10 | logLik: -1220147.96681948
#> Mon Nov 6 08:01:03 2023 .. Created module 11 | logLik: -1220818.37022325
#> Mon Nov 6 08:01:03 2023 .. Created module 12 | logLik: -1221489.07685946
#> Mon Nov 6 08:01:03 2023 .. Created module 13 | logLik: -1222032.53497571
#> Mon Nov 6 08:01:03 2023 .. Created module 14 | logLik: -1222712.17543857
#> Mon Nov 6 08:01:03 2023 .. Created module 15 | logLik: -1223268.97596756
#> Mon Nov 6 08:01:03 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive module splitting. Total time: 2.128508 secs
#> ==================================================
plotGridSearchPerplexity(moduleSplit)
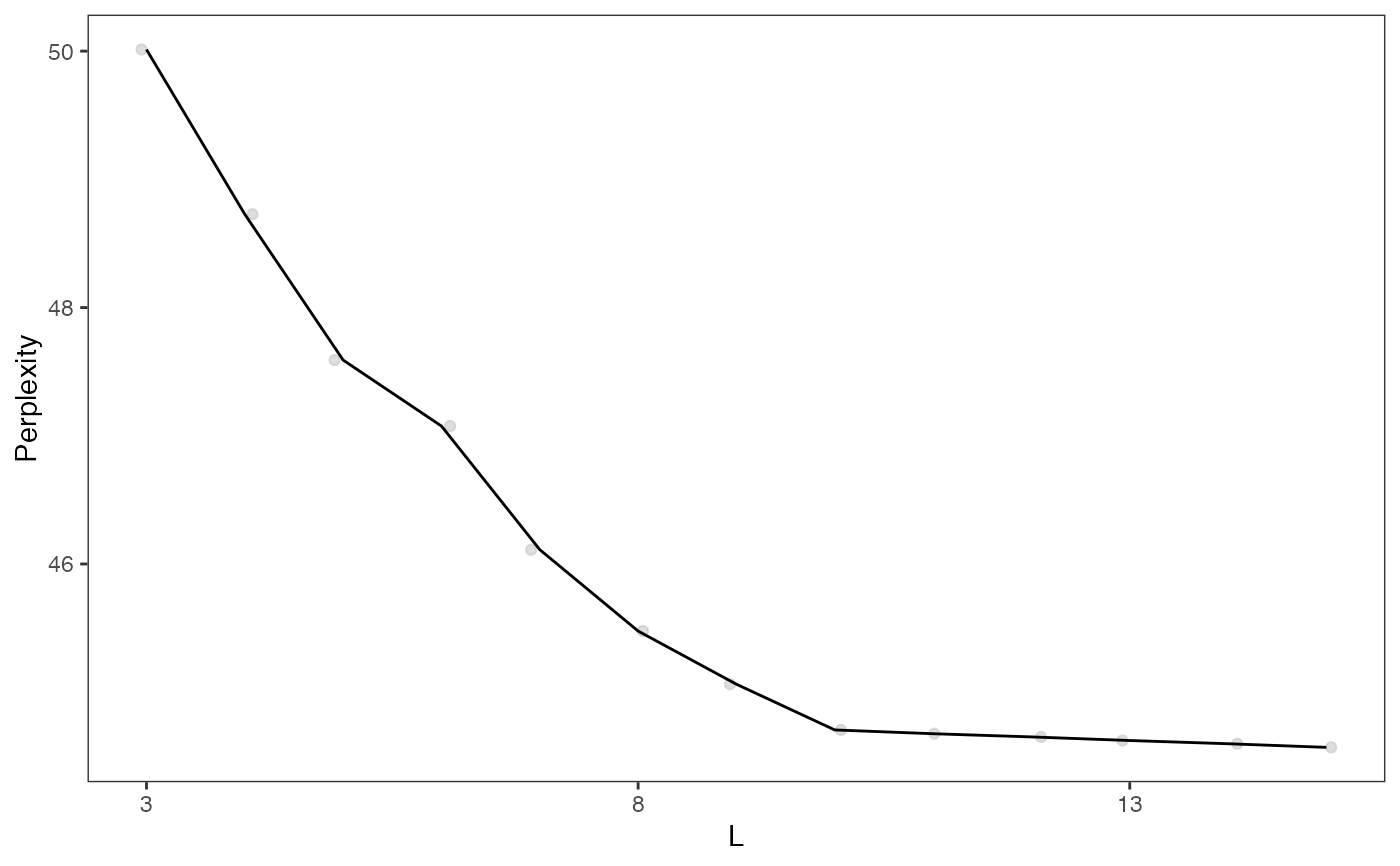 moduleSplitSelect <- subsetCeldaList(moduleSplit, list(L = 10))
## Then use module labels for initialization in \link{recursiveSplitCell} to
## produce \link{celda_CG} bi-clustering models
cellSplit <- recursiveSplitCell(sceCeldaCG,
initialK = 3, maxK = 7, yInit = celdaModules(moduleSplitSelect))
#> ==================================================
#> Starting recursive cell population splitting.
#> ==================================================
#> Mon Nov 6 08:01:04 2023 .. Collapsing to 10 modules
#> Mon Nov 6 08:01:04 2023 .. Initializing with 3 populations
#> Mon Nov 6 08:01:04 2023 .. Current cell population 4 | logLik: -1225286.49558716
#> Mon Nov 6 08:01:04 2023 .. Current cell population 5 | logLik: -1212955.15575681
#> Mon Nov 6 08:01:04 2023 .. Current cell population 6 | logLik: -1212982.74290613
#> Mon Nov 6 08:01:05 2023 .. Current cell population 7 | logLik: -1213005.40337891
#> Mon Nov 6 08:01:05 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive cell population splitting. Total time: 0.9962049 secs
#> ==================================================
plotGridSearchPerplexity(cellSplit)
moduleSplitSelect <- subsetCeldaList(moduleSplit, list(L = 10))
## Then use module labels for initialization in \link{recursiveSplitCell} to
## produce \link{celda_CG} bi-clustering models
cellSplit <- recursiveSplitCell(sceCeldaCG,
initialK = 3, maxK = 7, yInit = celdaModules(moduleSplitSelect))
#> ==================================================
#> Starting recursive cell population splitting.
#> ==================================================
#> Mon Nov 6 08:01:04 2023 .. Collapsing to 10 modules
#> Mon Nov 6 08:01:04 2023 .. Initializing with 3 populations
#> Mon Nov 6 08:01:04 2023 .. Current cell population 4 | logLik: -1225286.49558716
#> Mon Nov 6 08:01:04 2023 .. Current cell population 5 | logLik: -1212955.15575681
#> Mon Nov 6 08:01:04 2023 .. Current cell population 6 | logLik: -1212982.74290613
#> Mon Nov 6 08:01:05 2023 .. Current cell population 7 | logLik: -1213005.40337891
#> Mon Nov 6 08:01:05 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive cell population splitting. Total time: 0.9962049 secs
#> ==================================================
plotGridSearchPerplexity(cellSplit)
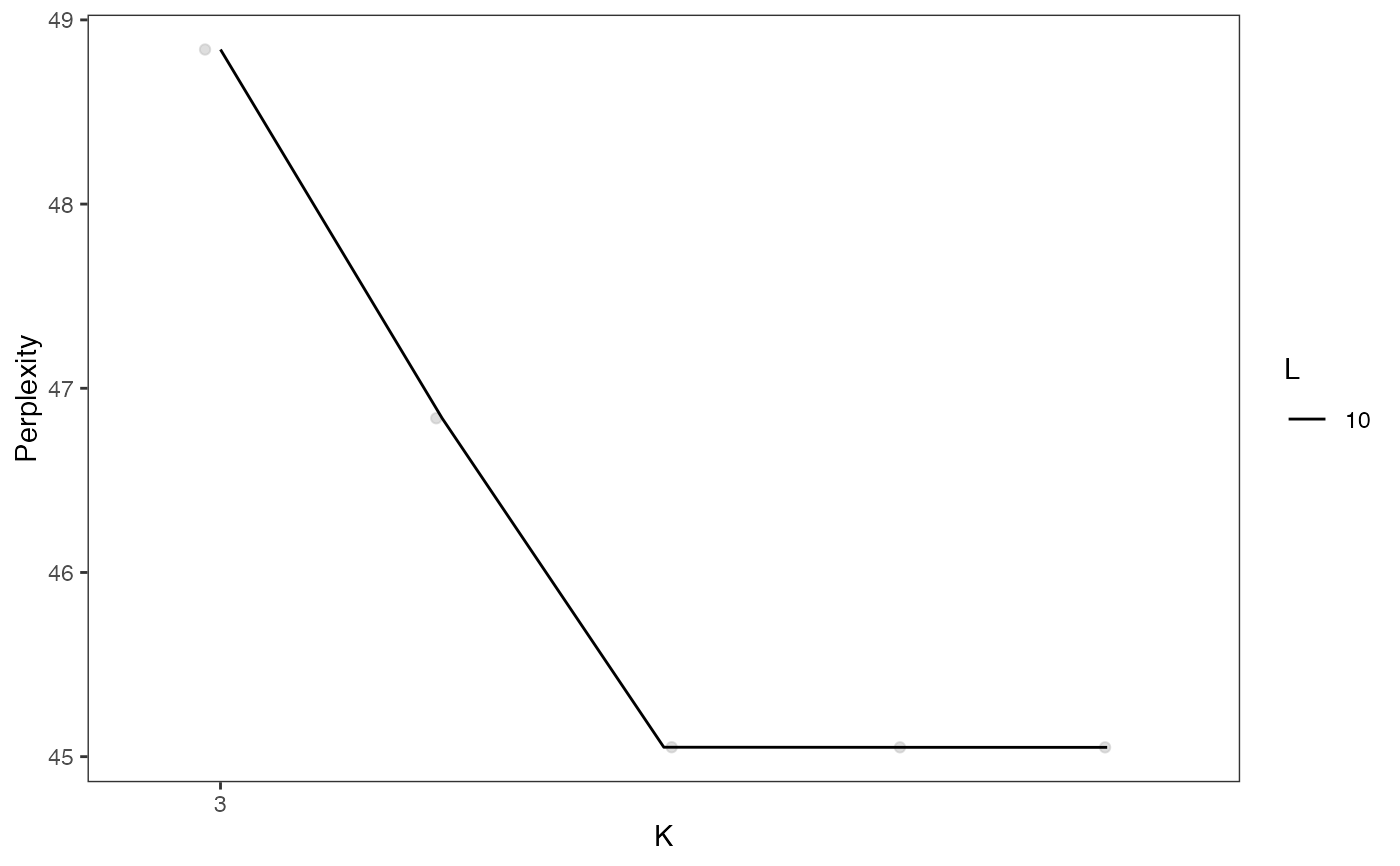 sce <- subsetCeldaList(cellSplit, list(K = 5, L = 10))
data(celdaCGSim, celdaCSim)
## Create models that range from K = 3 to K = 7 by recursively splitting
## cell populations into two to produce \link{celda_C} cell clustering models
sce <- recursiveSplitCell(celdaCSim$counts, initialK = 3, maxK = 7)
#> ==================================================
#> Starting recursive cell population splitting.
#> ==================================================
#> Mon Nov 6 08:01:05 2023 .. Initializing with 3 populations
#> Mon Nov 6 08:01:05 2023 .. Current cell population 4 | logLik: -1341630.1679001
#> Mon Nov 6 08:01:05 2023 .. Current cell population 5 | logLik: -1327506.91718317
#> Mon Nov 6 08:01:05 2023 .. Current cell population 6 | logLik: -1315227.54586167
#> Mon Nov 6 08:01:05 2023 .. Current cell population 7 | logLik: -1304393.65802293
#> Mon Nov 6 08:01:05 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive cell population splitting. Total time: 0.2063239 secs
#> ==================================================
## Alternatively, first identify features modules using
## \link{recursiveSplitModule}
moduleSplit <- recursiveSplitModule(celdaCGSim$counts,
initialL = 3, maxL = 15)
#> ==================================================
#> Starting recursive module splitting.
#> ==================================================
#> Mon Nov 6 08:01:06 2023 .. Collapsing to 100 temporary cell populations
#> Mon Nov 6 08:01:06 2023 .. Initializing with 3 modules
#> Mon Nov 6 08:01:07 2023 .. Created module 4 | logLik: -1243396.62348886
#> Mon Nov 6 08:01:07 2023 .. Created module 5 | logLik: -1237610.11790137
#> Mon Nov 6 08:01:07 2023 .. Created module 6 | logLik: -1232128.87013396
#> Mon Nov 6 08:01:07 2023 .. Created module 7 | logLik: -1227611.8250329
#> Mon Nov 6 08:01:07 2023 .. Created module 8 | logLik: -1225618.06184004
#> Mon Nov 6 08:01:07 2023 .. Created module 9 | logLik: -1223967.77531912
#> Mon Nov 6 08:01:07 2023 .. Created module 10 | logLik: -1222801.11395987
#> Mon Nov 6 08:01:07 2023 .. Created module 11 | logLik: -1223402.66903597
#> Mon Nov 6 08:01:07 2023 .. Created module 12 | logLik: -1224026.19892208
#> Mon Nov 6 08:01:07 2023 .. Created module 13 | logLik: -1224675.63005464
#> Mon Nov 6 08:01:07 2023 .. Created module 14 | logLik: -1225317.91966369
#> Mon Nov 6 08:01:07 2023 .. Created module 15 | logLik: -1225971.50555157
#> Mon Nov 6 08:01:07 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive module splitting. Total time: 1.948939 secs
#> ==================================================
plotGridSearchPerplexity(moduleSplit)
sce <- subsetCeldaList(cellSplit, list(K = 5, L = 10))
data(celdaCGSim, celdaCSim)
## Create models that range from K = 3 to K = 7 by recursively splitting
## cell populations into two to produce \link{celda_C} cell clustering models
sce <- recursiveSplitCell(celdaCSim$counts, initialK = 3, maxK = 7)
#> ==================================================
#> Starting recursive cell population splitting.
#> ==================================================
#> Mon Nov 6 08:01:05 2023 .. Initializing with 3 populations
#> Mon Nov 6 08:01:05 2023 .. Current cell population 4 | logLik: -1341630.1679001
#> Mon Nov 6 08:01:05 2023 .. Current cell population 5 | logLik: -1327506.91718317
#> Mon Nov 6 08:01:05 2023 .. Current cell population 6 | logLik: -1315227.54586167
#> Mon Nov 6 08:01:05 2023 .. Current cell population 7 | logLik: -1304393.65802293
#> Mon Nov 6 08:01:05 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive cell population splitting. Total time: 0.2063239 secs
#> ==================================================
## Alternatively, first identify features modules using
## \link{recursiveSplitModule}
moduleSplit <- recursiveSplitModule(celdaCGSim$counts,
initialL = 3, maxL = 15)
#> ==================================================
#> Starting recursive module splitting.
#> ==================================================
#> Mon Nov 6 08:01:06 2023 .. Collapsing to 100 temporary cell populations
#> Mon Nov 6 08:01:06 2023 .. Initializing with 3 modules
#> Mon Nov 6 08:01:07 2023 .. Created module 4 | logLik: -1243396.62348886
#> Mon Nov 6 08:01:07 2023 .. Created module 5 | logLik: -1237610.11790137
#> Mon Nov 6 08:01:07 2023 .. Created module 6 | logLik: -1232128.87013396
#> Mon Nov 6 08:01:07 2023 .. Created module 7 | logLik: -1227611.8250329
#> Mon Nov 6 08:01:07 2023 .. Created module 8 | logLik: -1225618.06184004
#> Mon Nov 6 08:01:07 2023 .. Created module 9 | logLik: -1223967.77531912
#> Mon Nov 6 08:01:07 2023 .. Created module 10 | logLik: -1222801.11395987
#> Mon Nov 6 08:01:07 2023 .. Created module 11 | logLik: -1223402.66903597
#> Mon Nov 6 08:01:07 2023 .. Created module 12 | logLik: -1224026.19892208
#> Mon Nov 6 08:01:07 2023 .. Created module 13 | logLik: -1224675.63005464
#> Mon Nov 6 08:01:07 2023 .. Created module 14 | logLik: -1225317.91966369
#> Mon Nov 6 08:01:07 2023 .. Created module 15 | logLik: -1225971.50555157
#> Mon Nov 6 08:01:07 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive module splitting. Total time: 1.948939 secs
#> ==================================================
plotGridSearchPerplexity(moduleSplit)
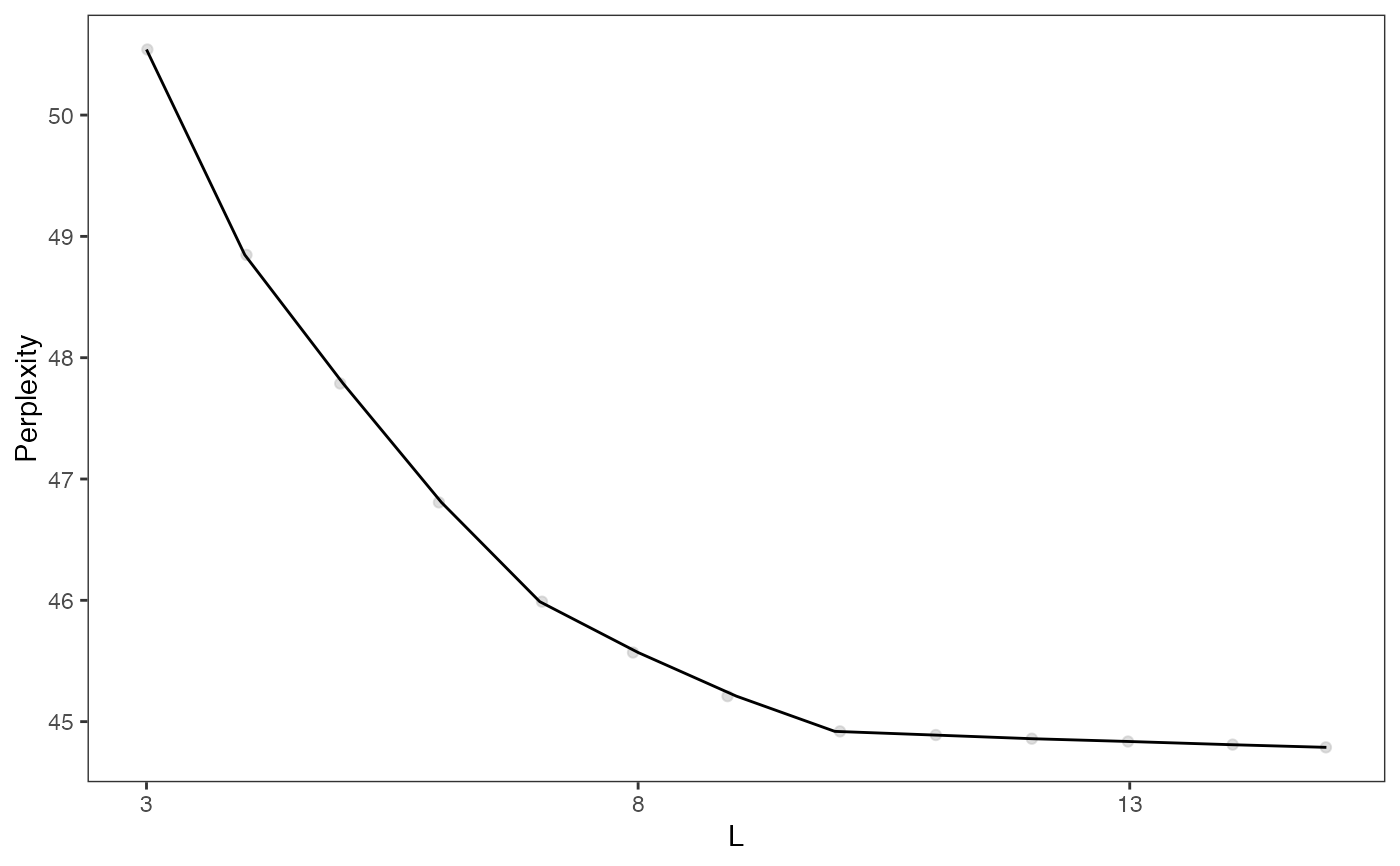 moduleSplitSelect <- subsetCeldaList(moduleSplit, list(L = 10))
## Then use module labels for initialization in \link{recursiveSplitCell} to
## produce \link{celda_CG} bi-clustering models
cellSplit <- recursiveSplitCell(celdaCGSim$counts,
initialK = 3, maxK = 7, yInit = celdaModules(moduleSplitSelect))
#> ==================================================
#> Starting recursive cell population splitting.
#> ==================================================
#> Mon Nov 6 08:01:08 2023 .. Collapsing to 10 modules
#> Mon Nov 6 08:01:08 2023 .. Initializing with 3 populations
#> Mon Nov 6 08:01:10 2023 .. Current cell population 4 | logLik: -1227944.5458832
#> Mon Nov 6 08:01:10 2023 .. Current cell population 5 | logLik: -1215605.08613503
#> Mon Nov 6 08:01:10 2023 .. Current cell population 6 | logLik: -1215627.62281773
#> Mon Nov 6 08:01:10 2023 .. Current cell population 7 | logLik: -1215651.32538066
#> Mon Nov 6 08:01:10 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive cell population splitting. Total time: 1.931995 secs
#> ==================================================
plotGridSearchPerplexity(cellSplit)
moduleSplitSelect <- subsetCeldaList(moduleSplit, list(L = 10))
## Then use module labels for initialization in \link{recursiveSplitCell} to
## produce \link{celda_CG} bi-clustering models
cellSplit <- recursiveSplitCell(celdaCGSim$counts,
initialK = 3, maxK = 7, yInit = celdaModules(moduleSplitSelect))
#> ==================================================
#> Starting recursive cell population splitting.
#> ==================================================
#> Mon Nov 6 08:01:08 2023 .. Collapsing to 10 modules
#> Mon Nov 6 08:01:08 2023 .. Initializing with 3 populations
#> Mon Nov 6 08:01:10 2023 .. Current cell population 4 | logLik: -1227944.5458832
#> Mon Nov 6 08:01:10 2023 .. Current cell population 5 | logLik: -1215605.08613503
#> Mon Nov 6 08:01:10 2023 .. Current cell population 6 | logLik: -1215627.62281773
#> Mon Nov 6 08:01:10 2023 .. Current cell population 7 | logLik: -1215651.32538066
#> Mon Nov 6 08:01:10 2023 .. Calculating perplexity
#> ==================================================
#> Completed recursive cell population splitting. Total time: 1.931995 secs
#> ==================================================
plotGridSearchPerplexity(cellSplit)
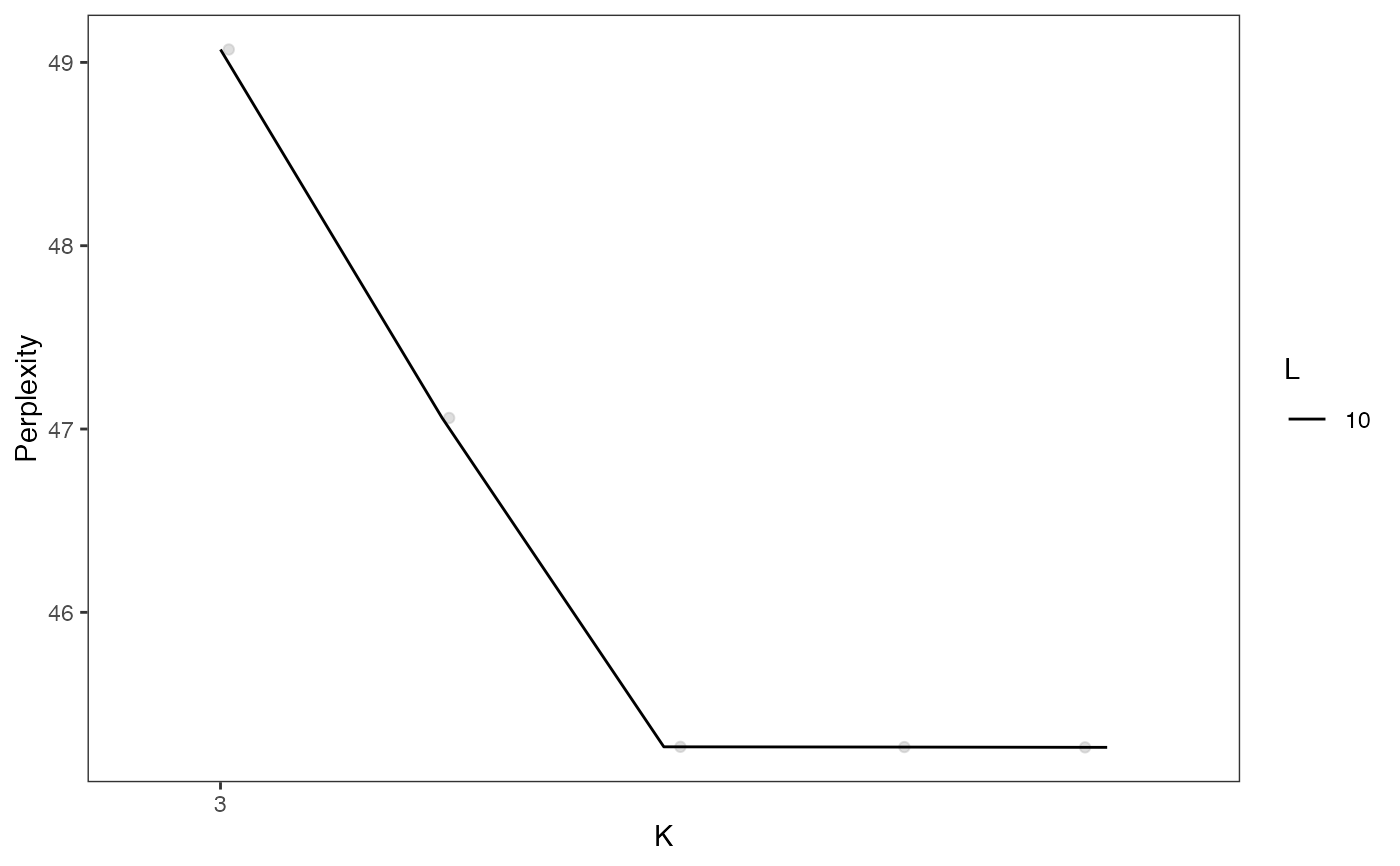 sce <- subsetCeldaList(cellSplit, list(K = 5, L = 10))
sce <- subsetCeldaList(cellSplit, list(K = 5, L = 10))