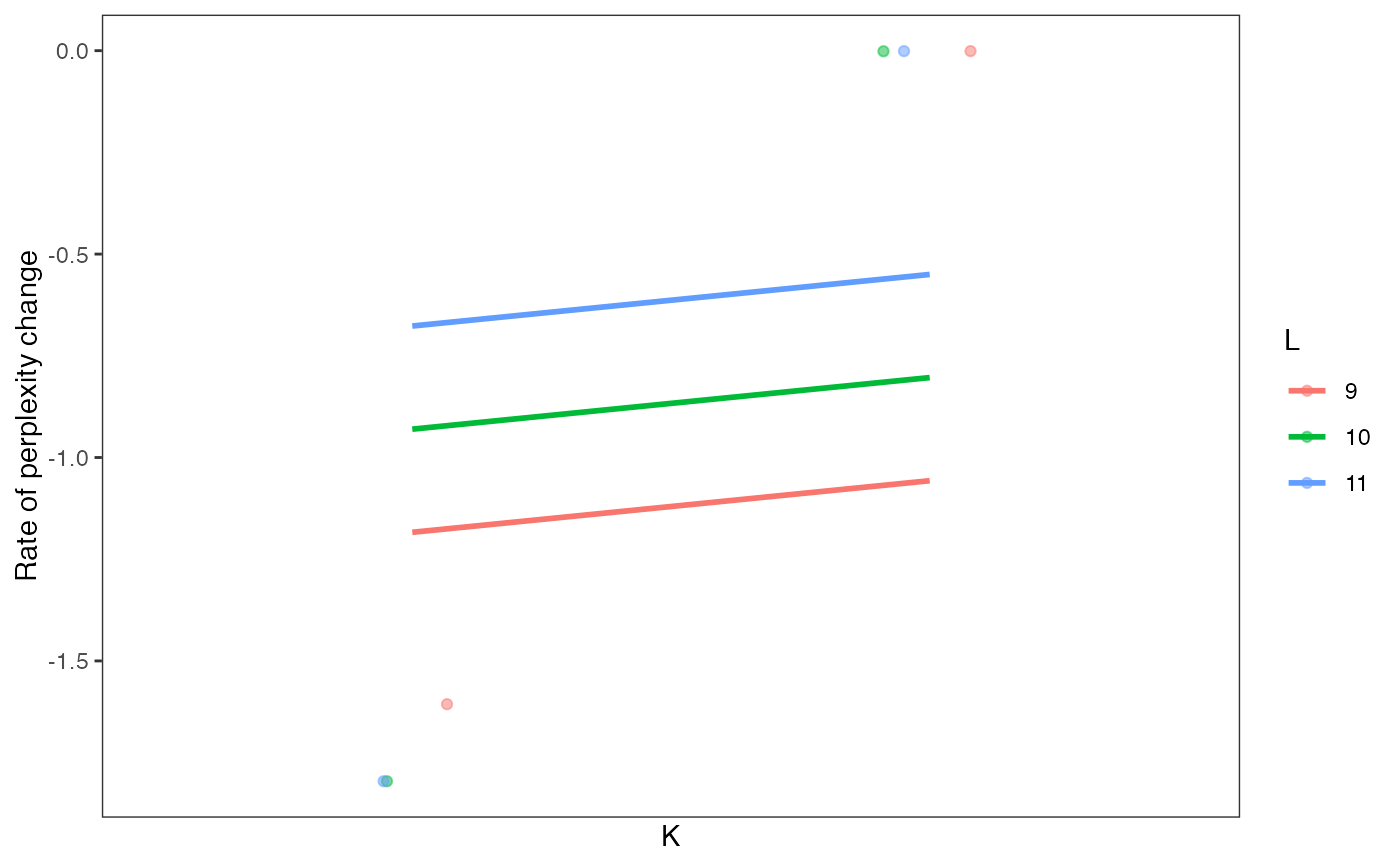
Visualize perplexity differences of every model in a celdaList, by unique K/L combinations.
plotRPC(x, altExpName = "featureSubset", sep = 5, alpha = 0.5)
# S4 method for SingleCellExperiment
plotRPC(x, altExpName = "featureSubset", sep = 5, alpha = 0.5)
# S4 method for celdaList
plotRPC(x, sep = 5, alpha = 0.5)Arguments
- x
Can be one of
A SingleCellExperiment object returned from
celdaGridSearch,recursiveSplitModule, orrecursiveSplitCell. Must contain a list named"celda_grid_search"inmetadata(x).celdaList object.
- altExpName
The name for the altExp slot to use. Default "featureSubset".
- sep
Numeric. Breaks in the x axis of the resulting plot.
- alpha
Numeric. Passed to geom_jitter. Opacity of the points. Values of alpha range from 0 to 1, with lower values corresponding to more transparent colors.
Value
A ggplot plot object showing perplexity differences as a function of clustering parameters.
Examples
data(sceCeldaCGGridSearch)
sce <- resamplePerplexity(sceCeldaCGGridSearch)
plotRPC(sce)
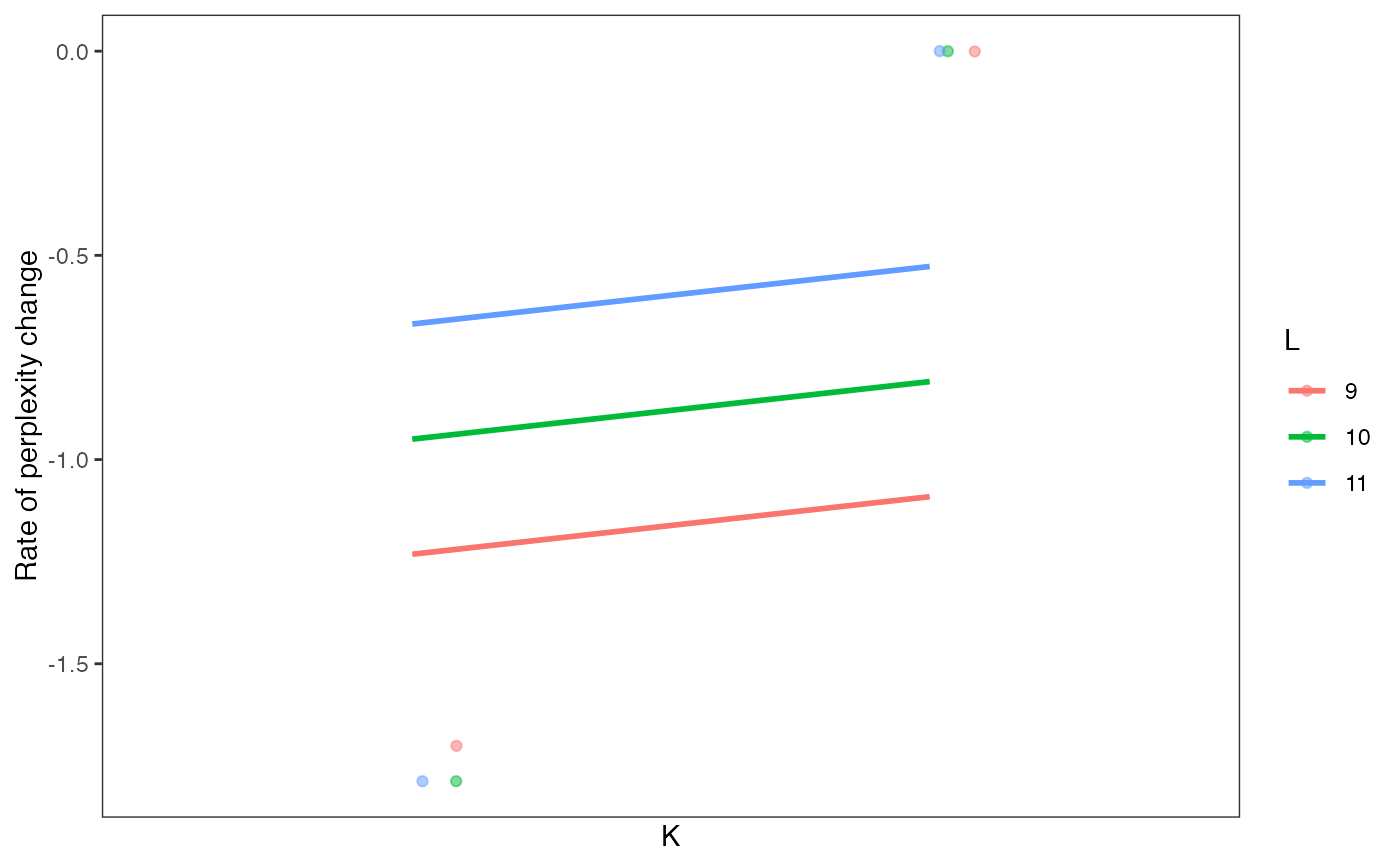 data(celdaCGSim, celdaCGGridSearchRes)
## Run various combinations of parameters with 'celdaGridSearch'
celdaCGGridSearchRes <- resamplePerplexity(
celdaCGSim$counts,
celdaCGGridSearchRes)
plotRPC(celdaCGGridSearchRes)
data(celdaCGSim, celdaCGGridSearchRes)
## Run various combinations of parameters with 'celdaGridSearch'
celdaCGGridSearchRes <- resamplePerplexity(
celdaCGSim$counts,
celdaCGGridSearchRes)
plotRPC(celdaCGGridSearchRes)