Identifies contamination from factors such as ambient RNA in single cell genomic datasets.
decontX(x, ...)
# S4 method for SingleCellExperiment
decontX(
x,
assayName = "counts",
z = NULL,
batch = NULL,
background = NULL,
bgAssayName = NULL,
bgBatch = NULL,
maxIter = 500,
delta = c(10, 10),
estimateDelta = TRUE,
convergence = 0.001,
iterLogLik = 10,
varGenes = 5000,
dbscanEps = 1,
seed = 12345,
logfile = NULL,
verbose = TRUE
)
# S4 method for ANY
decontX(
x,
z = NULL,
batch = NULL,
background = NULL,
bgBatch = NULL,
maxIter = 500,
delta = c(10, 10),
estimateDelta = TRUE,
convergence = 0.001,
iterLogLik = 10,
varGenes = 5000,
dbscanEps = 1,
seed = 12345,
logfile = NULL,
verbose = TRUE
)Arguments
- x
A numeric matrix of counts or a SingleCellExperiment with the matrix located in the assay slot under
assayName. Cells in each batch will be subsetted and converted to a sparse matrix of classdgCMatrixfrom package Matrix before analysis. This object should only contain filtered cells after cell calling. Empty cell barcodes (low expression droplets before cell calling) are not needed to run DecontX.- ...
For the generic, further arguments to pass to each method.
- assayName
Character. Name of the assay to use if
xis a SingleCellExperiment.- z
Numeric or character vector. Cell cluster labels. If NULL, PCA will be used to reduce the dimensionality of the dataset initially, 'umap' from the 'uwot' package will be used to further reduce the dataset to 2 dimenions and the 'dbscan' function from the 'dbscan' package will be used to identify clusters of broad cell types. Default NULL.
- batch
Numeric or character vector. Batch labels for cells. If batch labels are supplied, DecontX is run on cells from each batch separately. Cells run in different channels or assays should be considered different batches. Default NULL.
- background
A numeric matrix of counts or a SingleCellExperiment with the matrix located in the assay slot under
assayName. It should have the same data format asxexcept it contains the empty droplets instead of cells. When supplied, empirical distribution of transcripts from these empty droplets will be used as the contamination distribution. Default NULL.- bgAssayName
Character. Name of the assay to use if
backgroundis a SingleCellExperiment. Default to same asassayName.- bgBatch
Numeric or character vector. Batch labels for
background. Its unique values should be the same as those inbatch, such that each batch of cells have their corresponding batch of empty droplets as background, pointed by this parameter. Default to NULL.- maxIter
Integer. Maximum iterations of the EM algorithm. Default 500.
- delta
Numeric Vector of length 2. Concentration parameters for the Dirichlet prior for the contamination in each cell. The first element is the prior for the native counts while the second element is the prior for the contamination counts. These essentially act as pseudocounts for the native and contamination in each cell. If
estimateDelta = TRUE, this is only used to produce a random sample of proportions for an initial value of contamination in each cell. Thenfit_dirichletis used to updatedeltain each iteration. IfestimateDelta = FALSE, thendeltais fixed with these values for the entire inference procedure. Fixingdeltaand setting a high number in the second element will forcedecontXto be more aggressive and estimate higher levels of contamination at the expense of potentially removing native expression. Defaultc(10, 10).- estimateDelta
Boolean. Whether to update
deltaat each iteration.- convergence
Numeric. The EM algorithm will be stopped if the maximum difference in the contamination estimates between the previous and current iterations is less than this. Default 0.001.
- iterLogLik
Integer. Calculate log likelihood every
iterLogLikiteration. Default 10.- varGenes
Integer. The number of variable genes to use in dimensionality reduction before clustering. Variability is calcualted using
modelGeneVarfunction from the 'scran' package. Used only when z is not provided. Default 5000.- dbscanEps
Numeric. The clustering resolution parameter used in 'dbscan' to estimate broad cell clusters. Used only when z is not provided. Default 1.
- seed
Integer. Passed to with_seed. For reproducibility, a default value of 12345 is used. If NULL, no calls to with_seed are made.
- logfile
Character. Messages will be redirected to a file named `logfile`. If NULL, messages will be printed to stdout. Default NULL.
- verbose
Logical. Whether to print log messages. Default TRUE.
Value
If x is a matrix-like object, a list will be returned
with the following items:
decontXcounts:The decontaminated matrix. Values obtained from the variational inference procedure may be non-integer. However, integer counts can be obtained by rounding, e.g.
round(decontXcounts).contamination:Percentage of contamination in each cell.
estimates:List of estimated parameters for each batch. If z was not supplied, then the UMAP coordinates used to generated cell cluster labels will also be stored here.
z:Cell population/cluster labels used for analysis.
runParams:List of arguments used in the function call.
If x is a SingleCellExperiment, then the decontaminated
counts will be stored as an assay and can be accessed with
decontXcounts(x). The contamination values and cluster labels
will be stored in colData(x). estimates and runParams
will be stored in metadata(x)$decontX. The UMAPs used to generated
cell cluster labels will be stored in
reducedDims slot in x.
Examples
# Generate matrix with contamination
s <- simulateContamination(seed = 12345)
library(SingleCellExperiment)
sce <- SingleCellExperiment(list(counts = s$observedCounts))
sce <- decontX(sce)
#> --------------------------------------------------
#> Starting DecontX
#> --------------------------------------------------
#> Mon Nov 6 07:59:43 2023 .. Analyzing all cells
#> Mon Nov 6 07:59:43 2023 .... Converting to sparse matrix
#> Mon Nov 6 07:59:43 2023 .... Generating UMAP and estimating cell types
#> Mon Nov 6 07:59:47 2023 .... Estimating contamination
#> Mon Nov 6 07:59:47 2023 ...... Completed iteration: 9 | converge: 0.0009154
#> Mon Nov 6 07:59:47 2023 .. Calculating final decontaminated matrix
#> --------------------------------------------------
#> Completed DecontX. Total time: 3.348798 secs
#> --------------------------------------------------
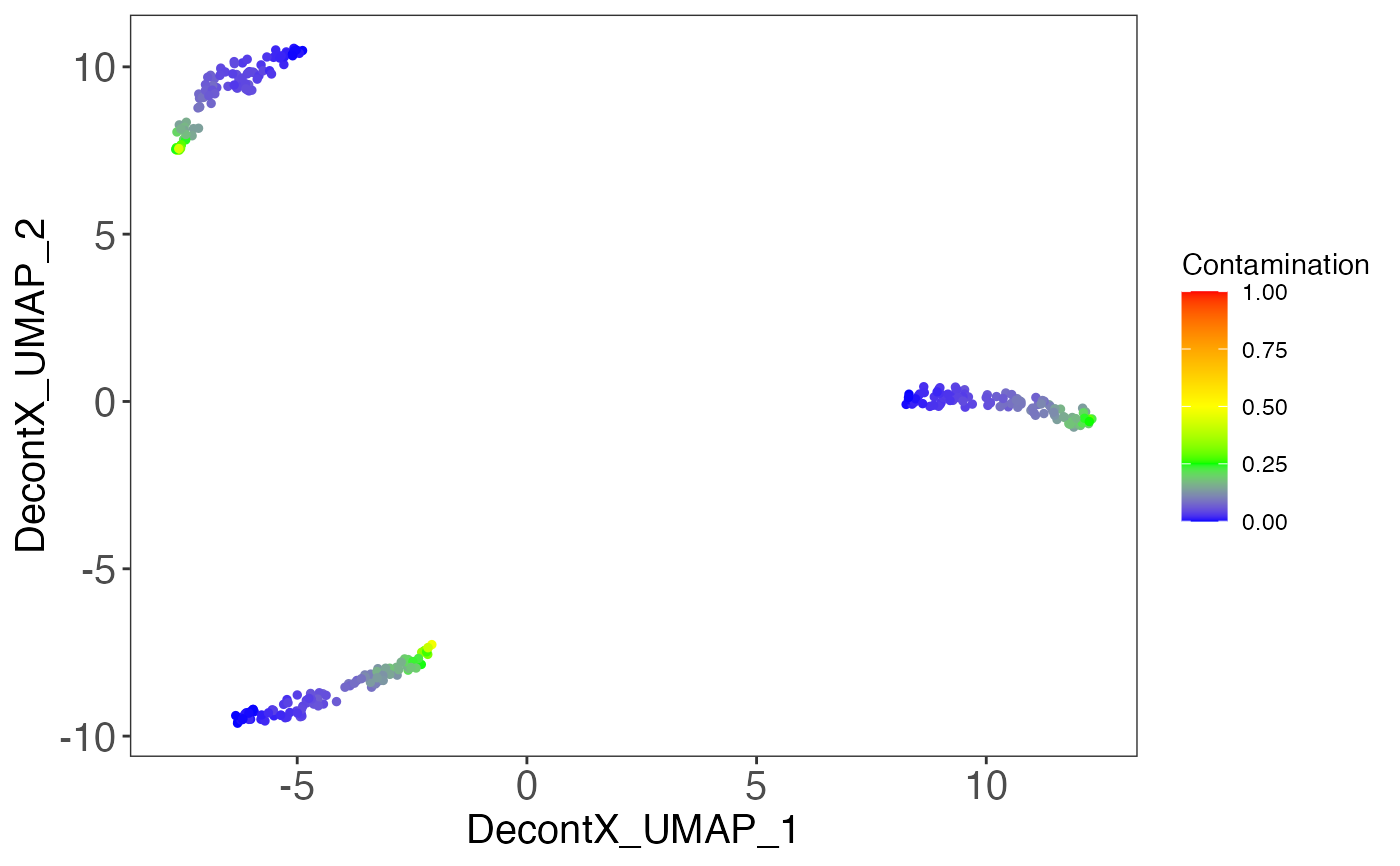
# Plot contamination on UMAP
plotDecontXContamination(sce)
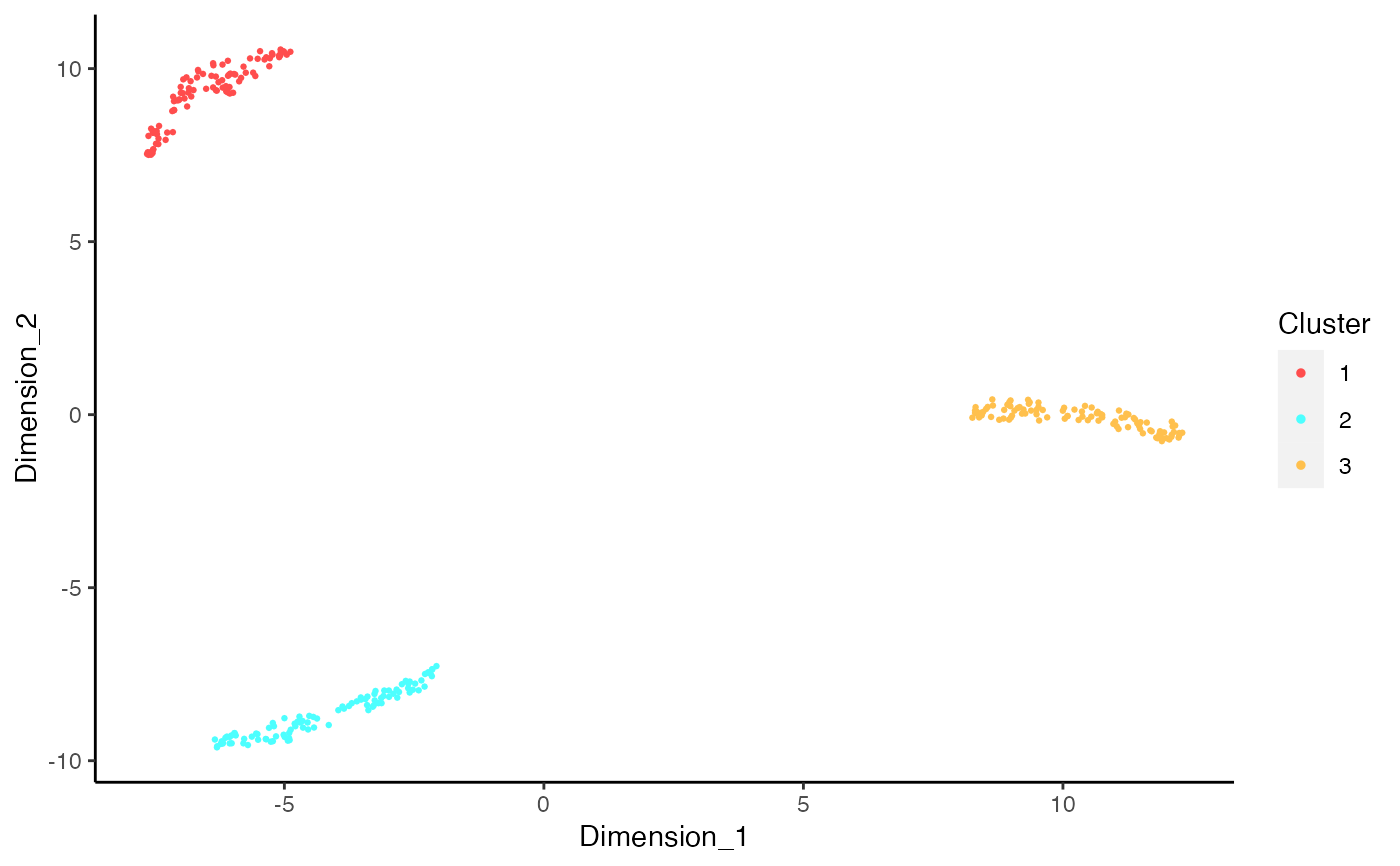
 # Plot decontX cluster labels
umap <- reducedDim(sce)
plotDimReduceCluster(x = sce$decontX_clusters,
dim1 = umap[, 1], dim2 = umap[, 2], )
# Plot decontX cluster labels
umap <- reducedDim(sce)
plotDimReduceCluster(x = sce$decontX_clusters,
dim1 = umap[, 1], dim2 = umap[, 2], )
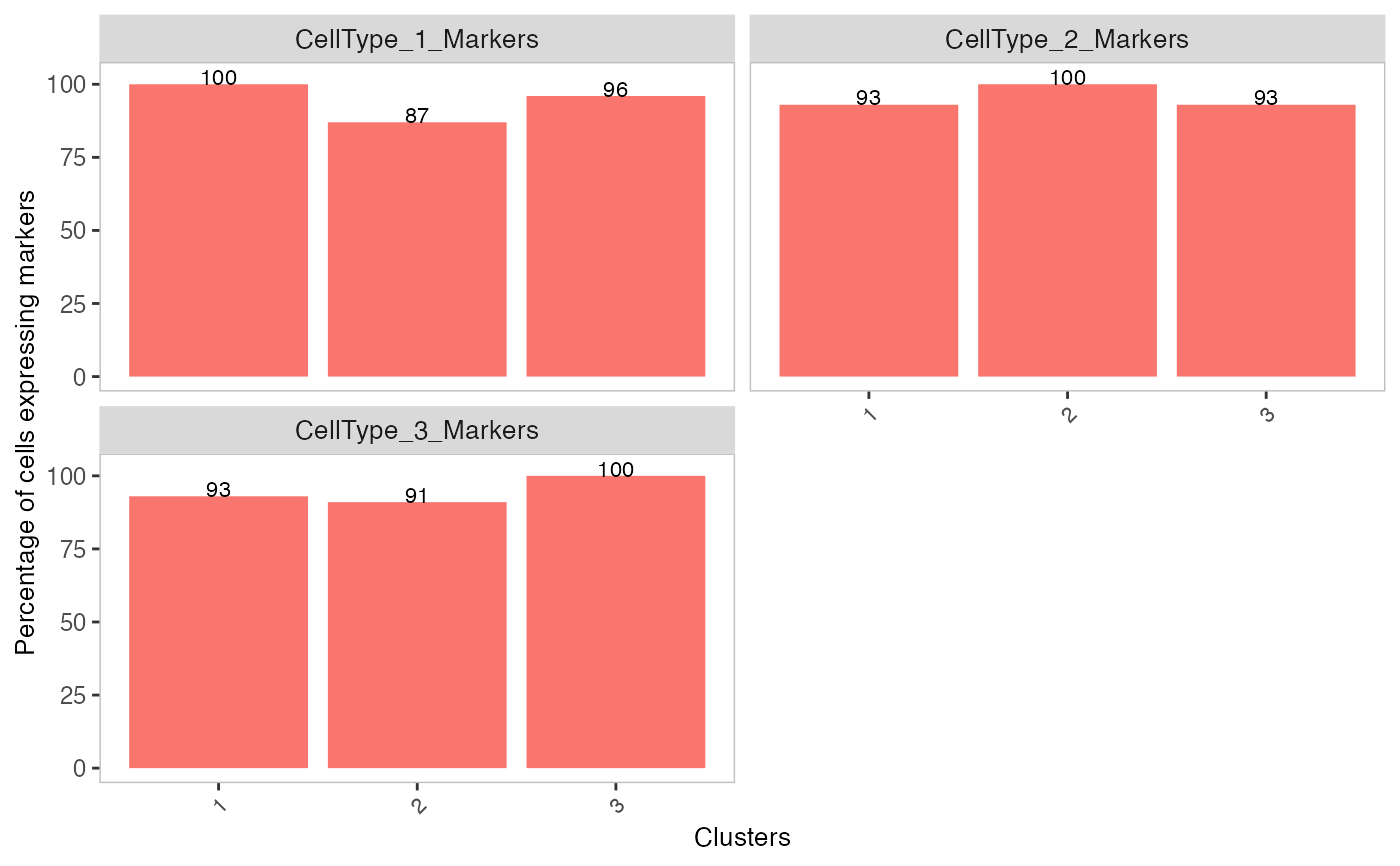
 # Plot percentage of marker genes detected
# in each cell cluster before decontamination
s$markers
#> $CellType_1_Markers
#> [1] "Gene_47" "Gene_32" "Gene_86"
#>
#> $CellType_2_Markers
#> [1] "Gene_70" "Gene_33" "Gene_48"
#>
#> $CellType_3_Markers
#> [1] "Gene_74" "Gene_26" "Gene_20"
#>
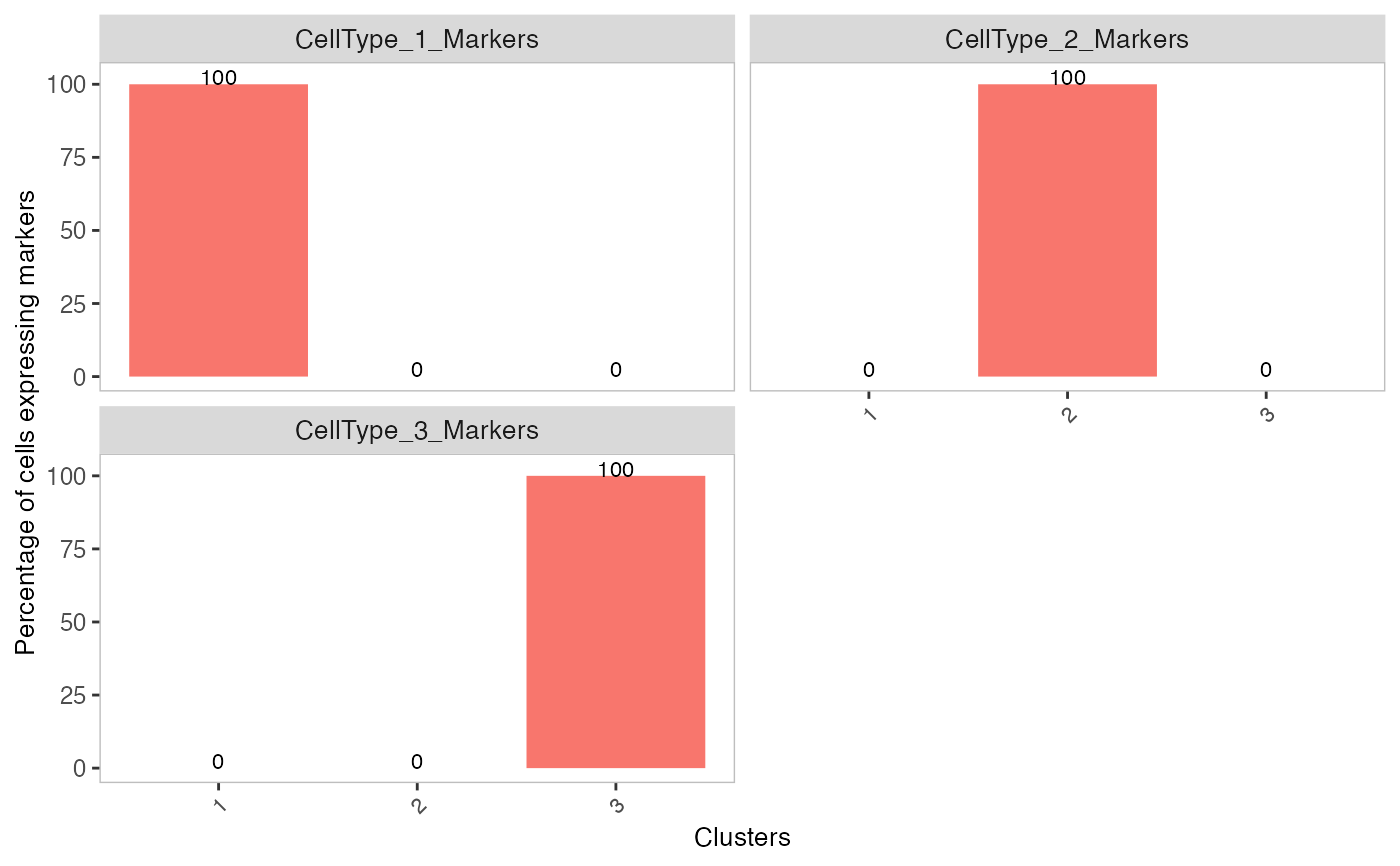
plotDecontXMarkerPercentage(sce, markers = s$markers, assayName = "counts")
# Plot percentage of marker genes detected
# in each cell cluster before decontamination
s$markers
#> $CellType_1_Markers
#> [1] "Gene_47" "Gene_32" "Gene_86"
#>
#> $CellType_2_Markers
#> [1] "Gene_70" "Gene_33" "Gene_48"
#>
#> $CellType_3_Markers
#> [1] "Gene_74" "Gene_26" "Gene_20"
#>
plotDecontXMarkerPercentage(sce, markers = s$markers, assayName = "counts")
 # Plot percentage of marker genes detected
# in each cell cluster after contamination
plotDecontXMarkerPercentage(sce, markers = s$markers,
assayName = "decontXcounts")
# Plot percentage of marker genes detected
# in each cell cluster after contamination
plotDecontXMarkerPercentage(sce, markers = s$markers,
assayName = "decontXcounts")
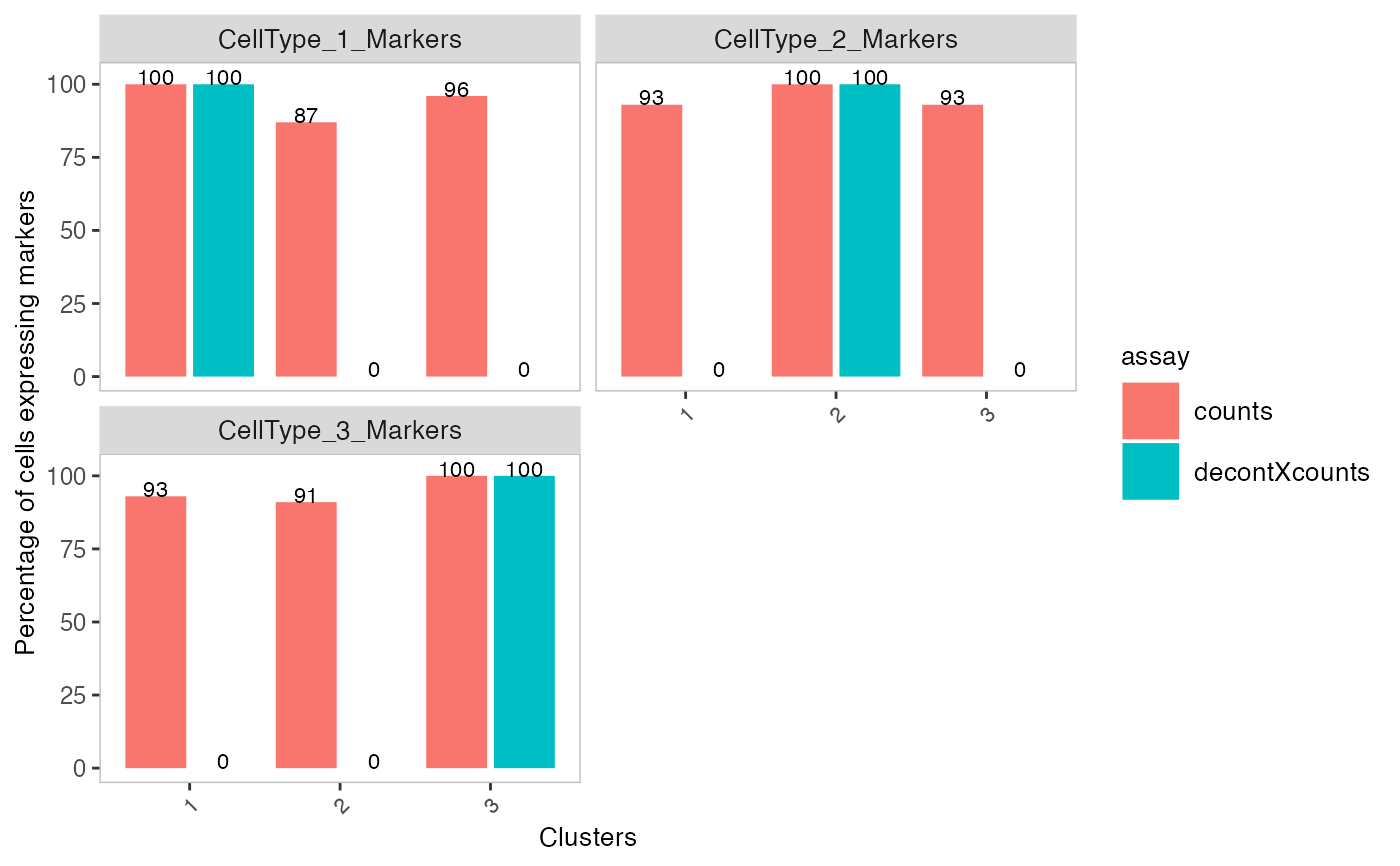
 # Plot percentage of marker genes detected in each cell
# comparing original and decontaminated counts side-by-side
plotDecontXMarkerPercentage(sce, markers = s$markers,
assayName = c("counts", "decontXcounts"))
# Plot percentage of marker genes detected in each cell
# comparing original and decontaminated counts side-by-side
plotDecontXMarkerPercentage(sce, markers = s$markers,
assayName = c("counts", "decontXcounts"))
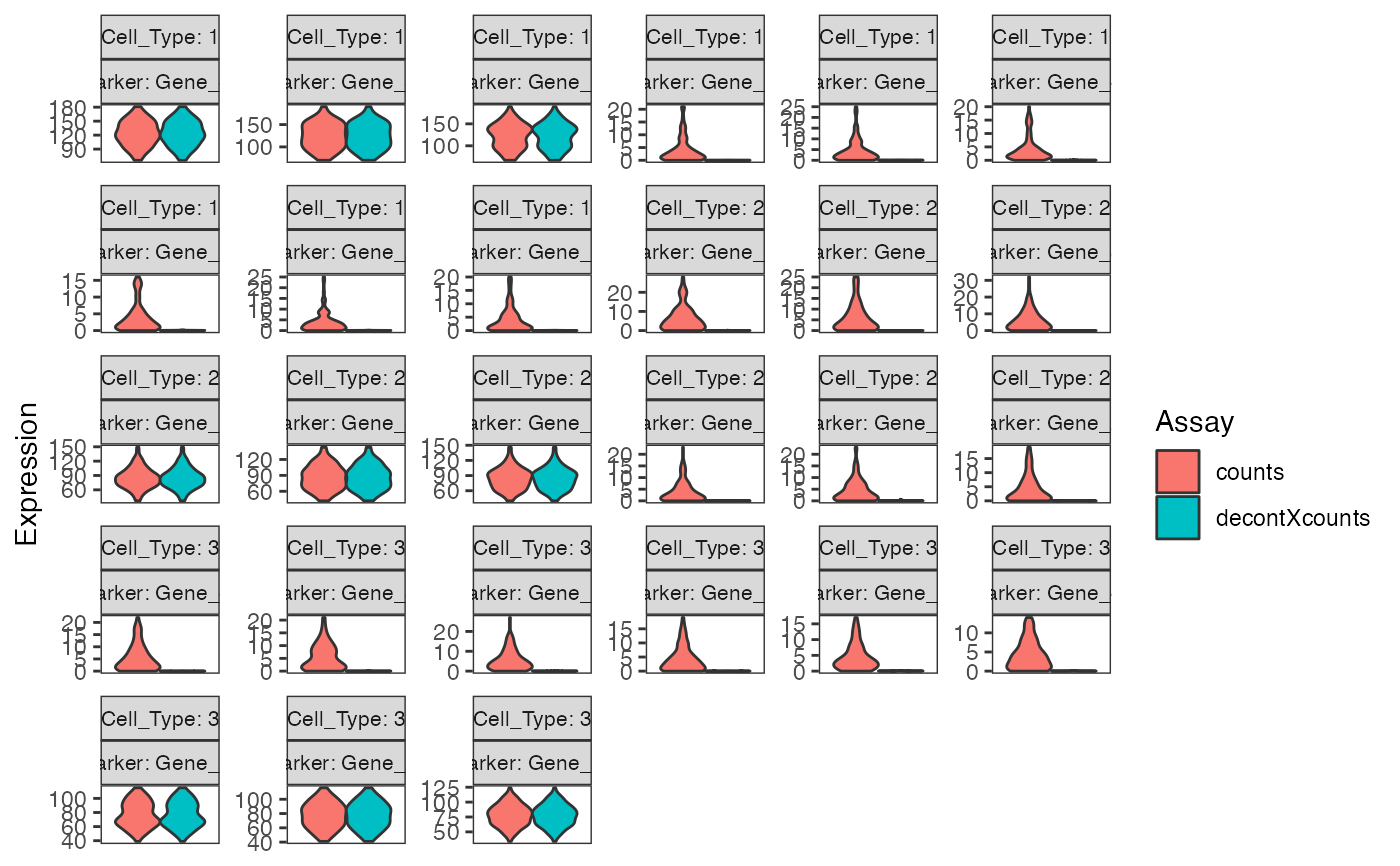
 # Plot raw counts of indiviual markers genes before
# and after decontamination
plotDecontXMarkerExpression(sce, unlist(s$markers))
# Plot raw counts of indiviual markers genes before
# and after decontamination
plotDecontXMarkerExpression(sce, unlist(s$markers))