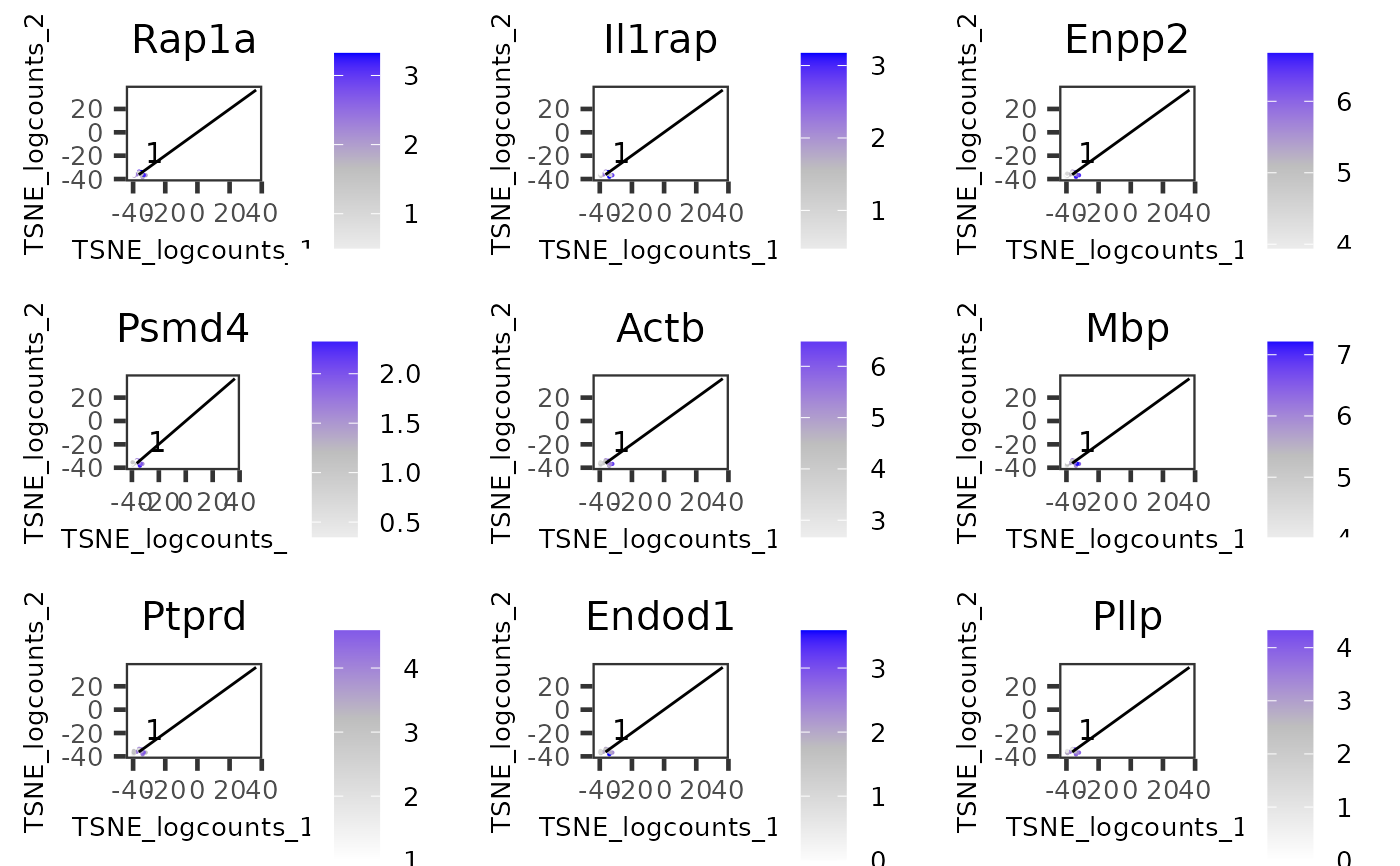
Plot features identified by runTSCANClusterDEAnalysis on
cell 2D embedding with MST overlaid
Source: R/runTSCAN.R
plotTSCANClusterDEG.RdA wrapper function which plot the top features expression
identified by runTSCANClusterDEAnalysis on the 2D embedding of
the cells cluster used in the analysis. The related MST edges are overlaid.
plotTSCANClusterDEG(
inSCE,
useCluster,
pathIndex = NULL,
useReducedDim = "UMAP",
topN = 9,
useAssay = NULL,
featureDisplay = metadata(inSCE)$featureDisplay,
combinePlot = c("all", "none")
)Arguments
- inSCE
Input SingleCellExperiment object.
- useCluster
Choose a cluster used for identifying DEG with
runTSCANClusterDEAnalysis. Required.- pathIndex
Specifies one of the branching paths from
useClusterand plot the top DEGs on this path. Ususally presented by the terminal cluster of a path. By defaultNULLplot top DEGs of all paths.- useReducedDim
A single character for the matrix of 2D embedding. Should exist in
reducedDimsslot. Default"UMAP".- topN
Integer. Use top N genes identified. Default
9.- useAssay
A single character for the feature expression matrix. Should exist in
assayNames(inSCE). DefaultNULLfor using the one used inrunTSCANClusterDEAnalysis.- featureDisplay
Specify the feature ID type to display. Users can set default value with
setSCTKDisplayRow.NULLor"rownames"specifies the rownames ofinSCE. Other character values indicatesrowDatavariable.- combinePlot
Must be either
"all"or"none"."all"will combine plots of each feature into a single.ggplotobject, while"none"will output a list of plots. Default"all".
Value
A .ggplot object of cell scatter plot, colored by the
expression of a gene identified by runTSCANClusterDEAnalysis,
with the layer of trajectory.
Examples
data("mouseBrainSubsetSCE", package = "singleCellTK")
mouseBrainSubsetSCE <- runTSCAN(inSCE = mouseBrainSubsetSCE,
useReducedDim = "PCA_logcounts")
#> Sat Mar 18 10:30:05 2023 ... Running 'scran SNN clustering' with 'louvain' algorithm
#> Sat Mar 18 10:30:06 2023 ... Identified 2 clusters
#> Sat Mar 18 10:30:06 2023 ... Running TSCAN to estimate pseudotime
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'
#> Also defined by ‘spam’
#> Sat Mar 18 10:30:06 2023 ... Clusters involved in path index 2 are: 1, 2
#> Sat Mar 18 10:30:06 2023 ... Number of estimated paths is 1
mouseBrainSubsetSCE <- runTSCANClusterDEAnalysis(inSCE = mouseBrainSubsetSCE,
useCluster = 1)
#> Sat Mar 18 10:30:06 2023 ... Finding DEG between TSCAN branches
#> Sat Mar 18 10:30:06 2023 ... Clusters involved in path index 2 are: 1, 2
plotTSCANClusterDEG(mouseBrainSubsetSCE, useCluster = 1,
useReducedDim = "TSNE_logcounts")