Yusuke Koga, Hanbing Song, Zachary R. Chalmers, Justin Newberg, Eejung Kim, Jian Carrot-Zhang, Daphnee Piou, Paz Polak, Sarki A. Abdulkadir, Elad Ziv, Matthew Meyerson, Garrett M. Frampton, Joshua D. Campbell and Franklin W. Huang.
Abstract
Purpose: African American (AFR) men have the highest mortality rate from prostate cancer (PCa) compared with men of other racial/ancestral groups. Differences in the spectrum of somatic genome alterations in tumors between AFR men and other populations have not been well-characterized due to a lack of inclusion of significant numbers in genomic studies.
Experimental design: To identify genomic alterations associated with race, we compared the frequencies of somatic alterations in PCa obtained from four publicly available datasets comprising 250 AFR and 611 European American (EUR) men and a targeted sequencing dataset from a commercial platform of 436 AFR and 3018 EUR men.
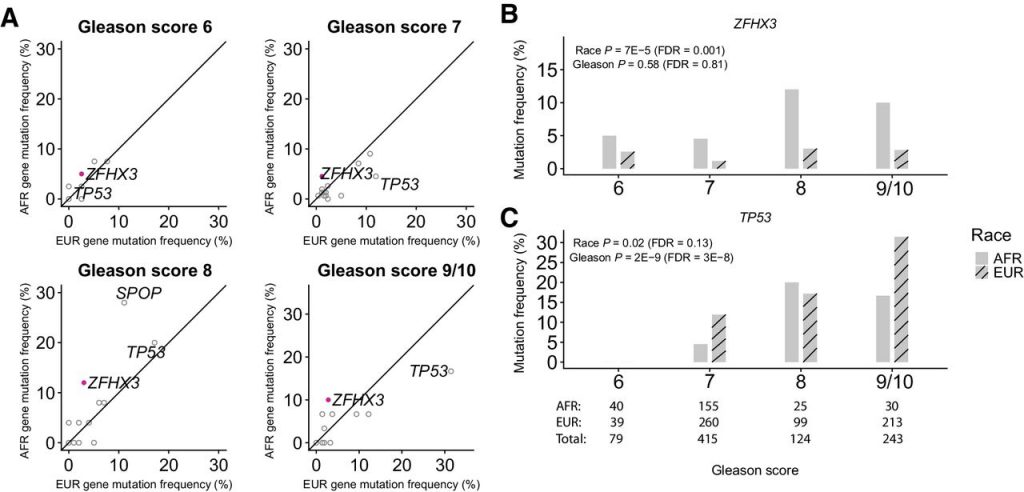
Results: Mutations in ZFHX3 as well as focal deletions in ETV3 were more frequent in tumors from AFR men. TP53 mutations were associated with increasing Gleason score. MYC amplifications were more frequent in tumors from AFR men with metastatic PCa, whereas deletions in PTEN and rearrangements in TMPRSS2-ERG were less frequent in tumors from AFR men. KMT2D truncations and CCND1 amplifications were more frequent in primary PCa from AFR men. Genomic features that could impact clinical decision making were not significantly different between the two groups including tumor mutation burden, MSI status, and genomic alterations in select DNA repair genes, CDK12, and in AR.
Conclusions: Although we identified some novel differences in AFR men compared with other populations, the frequencies of genomic alterations in current therapeutic targets for PCa were similar between AFR and EUR men, suggesting that existing precision medicine approaches could be equally beneficial if applied equitably.